Data preparation with dbt and BigQuery
Raw incoming data needs to go through a series of data preparation steps before it can be used for analysis. These steps include tasks such as type casting, renaming columns, cleaning values and identifying duplicates. Writing code to perform these tasks can become cumbersome if you have many data sources, and you end up with lots of repetitive code because almost the same pattern applies across data sources.
In this blog post, I will show you how to perform these common data preparation steps with data build tool (dbt) and BigQuery. At its core, dbt is a template compiler and runner that provides abstractions to easily write data pipelines in SQL. It focuses on the transform step in extract, load, transform (ELT), so it doesn’t extract or load data. If you are not familiar with dbt, I suggest that you start with this short introduction by Tristan Handy, the founder of dbt Labs. On the other hand, BigQuery is a modern cloud data warehouse that offers high performance and scalability in a serverless mode.
Source data
The source data comes from the legacy release of the USDA National Nutrient Database for Standard Reference. It is a set of CSV files that contain food composition data in the United States. For this demo, I loaded four of the CSV files into a BigQuery “landing” dataset using the python script below:
# bigquery_landing_load.py
from google.cloud import bigquery
def load_csv_file(file_path, project_id, dataset_id, table_id):
client = bigquery.Client(project=project_id, location='US')
table_ref = client.dataset(dataset_id).table(table_id)
job_config = bigquery.LoadJobConfig()
job_config.source_format = bigquery.SourceFormat.CSV
job_config.field_delimiter = '^'
job_config.quote_character = '~'
job_config.write_disposition = bigquery.WriteDisposition.WRITE_TRUNCATE
with open(file_path, 'rb') as source_file:
job = client.load_table_from_file(source_file, table_ref, job_config=job_config)
job.result() # Wait for the load to complete
print("Loaded {} rows into {}:{}.".format(job.output_rows, dataset_id, table_id))
if __name__ == '__main__':
load_csv_file('data/SR-Leg_ASC/FOOD_DES.txt', 'bigquery-sandbox', 'dbt_demo_ldg', 'stdref_fd_desc')
load_csv_file('data/SR-Leg_ASC/FD_GROUP.txt', 'bigquery-sandbox', 'dbt_demo_ldg', 'stdref_fd_group')
load_csv_file('data/SR-Leg_ASC/NUT_DATA.txt', 'bigquery-sandbox', 'dbt_demo_ldg', 'stdref_nut_data')
load_csv_file('data/SR-Leg_ASC/NUTR_DEF.txt', 'bigquery-sandbox', 'dbt_demo_ldg', 'stdref_nutr_def')
BigQuery datasets
The dbt_demo_ldg dataset contains the raw data loaded by the python script. All columns have the string data type since all data preparation steps will be performed later in dbt.
The dbt_demo_stg dataset contains views generated by dbt from staging models. These views reference tables in dbt_demo_ldg and perform all data preparation steps using SQL.
The dbt_demo_dwh dataset contains tables materialized by dbt from data warehouse models. These models reference staging models and apply specific business logic to prepare the data for consumption by end users.
dbt project
The image below shows the structure of the project. Here I’ll focus on the staging models and macros, since all data preparation steps happen there.
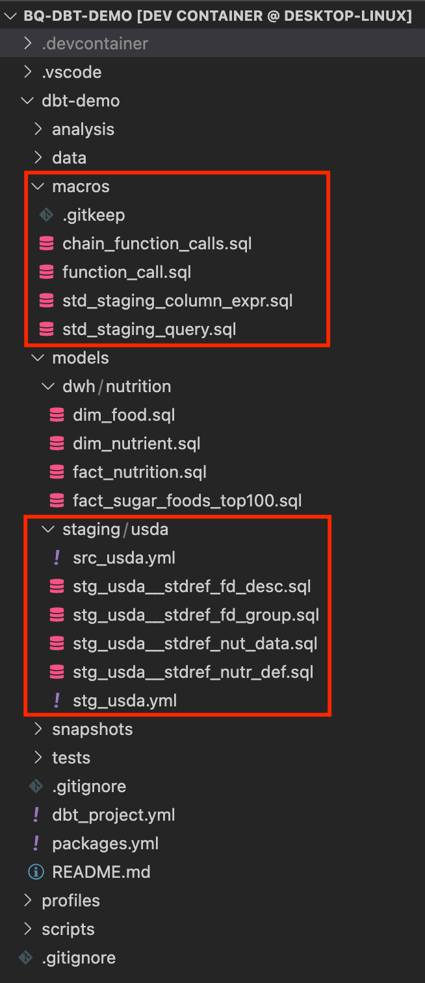
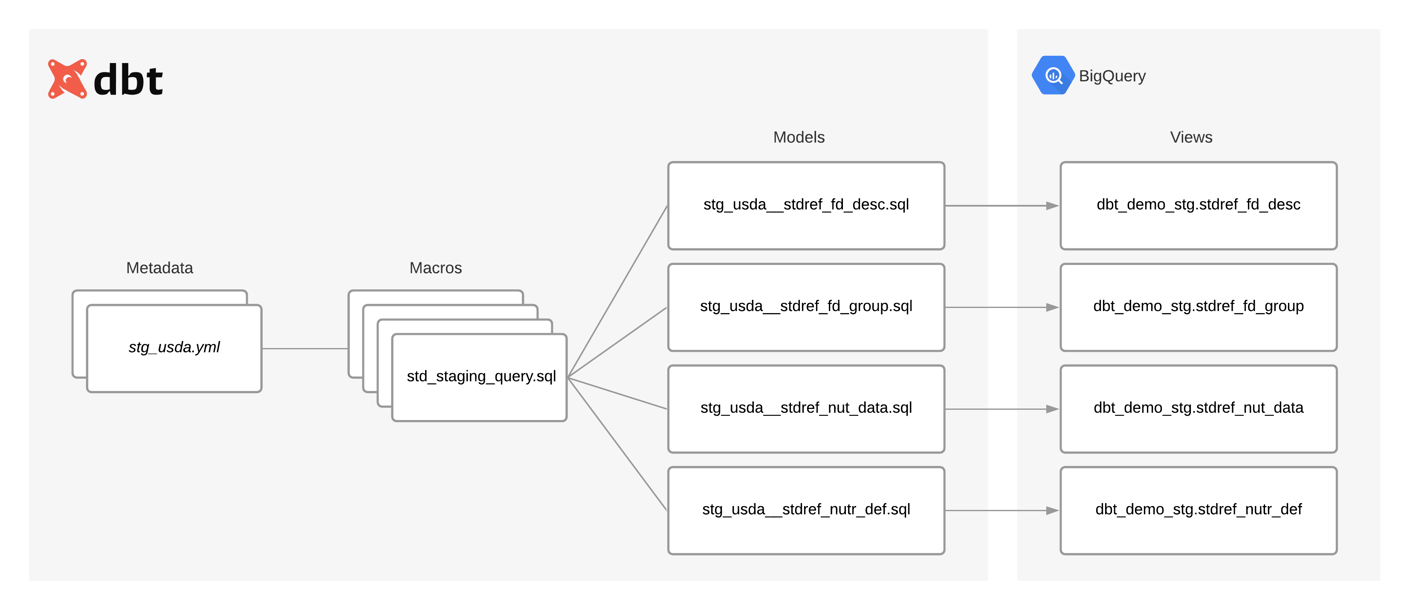
The following diagram shows the relationship between metadata, macros and models. It also shows how models materialize as views during deployment.
Metadata
The src_usda.yml file simply declares the tables that are available in the dbt_demo_ldg dataset. Models can then reference these tables using the source function:
# src_usda.yml
version: 2
sources:
- name: usda
schema: dbt_demo_ldg
tables:
- name: stdref_fd_desc
- name: stdref_fd_group
- name: stdref_nut_data
- name: stdref_nutr_def
The stg_usda.yml file contains the metadata for each model. This metadata defines the transformations that will be applied to the source data:
# stg_usda.yml
version: 2
models:
- name: stg_usda__stdref_fd_desc
columns:
- name: ndb_no
tests:
- unique
- not_null
meta:
data_type: string
- name: fdgrp_cd
meta:
data_type: string
- name: long_desc
meta:
data_type: string
- name: shrt_desc
meta:
data_type: string
- name: com_name
meta:
data_type: string
cleaning:
- ["trim"]
- ["nullif", "''"]
- name: manufac_name
meta:
data_type: string
cleaning:
- ["trim"]
- ["nullif", "''"]
- name: survey
meta:
data_type: string
cleaning:
- ["trim"]
- ["nullif", "''"]
- name: ref_desc
meta:
data_type: string
cleaning:
- ["trim"]
- ["nullif", "''"]
- name: refuse
meta:
data_type: numeric
- name: sci_name
meta:
data_type: string
cleaning:
- ["trim"]
- ["nullif", "''"]
- name: n_factor
meta:
data_type: numeric
- name: pro_factor
meta:
data_type: numeric
- name: fat_factor
meta:
data_type: numeric
- name: cho_factor
meta:
data_type: numeric
- name: stg_usda__stdref_fd_group
meta:
uniqueness:
partition_by:
- fdgrp_code
order_by:
- "fdgrp_code asc"
columns:
- name: fdgrp_code
meta:
source_column: fdgrp_cd
data_type: string
cleaning:
- ["left", 4]
- name: fdgrp_desc
meta:
data_type: string
cleaning:
- ["left", 60]
- name: stg_usda__stdref_nut_data
tests:
- dbt_utils.unique_combination_of_columns:
combination_of_columns:
- ndb_no
- nutr_no
columns:
- name: ndb_no
meta:
data_type: string
- name: nutr_no
meta:
data_type: string
- name: nutr_val
meta:
data_type: numeric
- name: num_data_pts
meta:
data_type: numeric
- name: std_error
meta:
data_type: string
- name: src_cd
meta:
data_type: string
- name: deriv_cd
meta:
data_type: string
- name: ref_ndb_no
meta:
data_type: string
- name: add_nutr_mark
meta:
data_type: string
- name: num_studies
meta:
data_type: numeric
- name: min
meta:
data_type: numeric
- name: max
meta:
data_type: numeric
- name: df
meta:
data_type: numeric
- name: low_eb
meta:
data_type: numeric
- name: up_eb
meta:
data_type: numeric
- name: stat_cmt
meta:
data_type: string
- name: addmod_date
meta:
data_type: date
cleaning:
- ["safe.parse_date", "'%m/%d/%Y'"]
- name: stg_usda__stdref_nutr_def
columns:
- name: nutr_no
tests:
- unique
- not_null
meta:
data_type: string
- name: units
meta:
data_type: string
- name: tag_name
meta:
data_type: string
- name: nutr_desc
meta:
data_type: string
- name: num_dec
meta:
data_type: string
- name: sr_order
meta:
data_type: numeric
Type casting
The data_type key in each column specifies the data type that the macros will use when casting the source column. For each column, an _is_valid column is added to indicate if the casting could be performed without errors.
Renaming columns
The name key in each column specifies the name for the column alias in the generated code. The macros assume that the source column has the same name. To use different names, the source column name needs to be specified in the source_column key.
Cleaning values
The cleaning key specifies the list of functions to use for cleaning values in a column. The macros chain together those functions in the generated code filling in the name of the column. Each function needs to be specified as an array. The first element is the name of the SQL function, and the rest are its arguments.
Identifying duplicates
The uniqueness key at the model level tells the macros to add two additional columns in the generated code. The is_duplicate column helps identify duplicate values, and the row_num column assigns a row number to each duplicate value. The partition_by and order_by keys specify the list of columns that should be used by the windowing functions.
Models
The staging models are very simple: They all call the std_staging_query macro passing as parameters the source to use and the current model. That’s it! The macro will generate the code specific to the model. Whenever a model requires complex data preparation steps, specific code can be written instead of calling the macro:
# stg_usda__stdref_fd_desc.sql
with source as (
)
select
*
from source
# stg_usda__stdref_fd_group.sql
with source as (
)
select
*
from source
# stg_usda__stdref_nut_data.sql
with source as (
)
select
*
from source
# stg_usda__stdref_nutr_def.sql
with source as (
)
select
*
from source
Macros
The main macro is std_staging_query. It reads the metadata for a staging model and generates SQL code based on it:
# std_staging_query.sql
The std_staging_column_expr macro returns the code that processes a column (type casting and cleaning).
# std_staging_column_expr.sql
The function_call and chain_function_calls macros chain together the list of functions that clean values in a column:
# function_call.sql
# chain_function_calls.sql
Deployment
The last step is to deploy the dbt project to BigQuery by executing the dbt run command. First, it will compile the templates, then it will open a connection to BigQuery to materialize the models (as views or tables). The type of materialization depends on the configuration specified in the dbt_project.yml file:
$ dbt run --profiles-dir=../profiles/ Running with dbt=0.19.1 Found 8 models, 5 tests, 0 snapshots, 0 analyses, 342 macros, 0 operations, 0 seed files, 4 sources, 0 exposures 14:02:50 | Concurrency: 4 threads (target='dev') 14:02:50 | 14:02:50 | 4 of 8 START view model dbt_demo_stg.stdref_nutr_def................. [RUN] 14:02:50 | 2 of 8 START view model dbt_demo_stg.stdref_fd_group................. [RUN] 14:02:50 | 3 of 8 START view model dbt_demo_stg.stdref_nut_data................. [RUN] 14:02:50 | 1 of 8 START view model dbt_demo_stg.stdref_fd_desc.................. [RUN] 14:02:51 | 1 of 8 OK created view model dbt_demo_stg.stdref_fd_desc............. [OK in 0.91s] 14:02:51 | 2 of 8 OK created view model dbt_demo_stg.stdref_fd_group............ [OK in 0.94s] 14:02:51 | 5 of 8 START table model dbt_demo_dwh.dim_food....................... [RUN] 14:02:51 | 4 of 8 OK created view model dbt_demo_stg.stdref_nutr_def............ [OK in 0.98s] 14:02:51 | 6 of 8 START table model dbt_demo_dwh.dim_nutrient................... [RUN] 14:02:51 | 3 of 8 OK created view model dbt_demo_stg.stdref_nut_data............ [OK in 1.08s] 14:02:51 | 7 of 8 START table model dbt_demo_dwh.fact_nutrition................. [RUN] 14:02:54 | 6 of 8 OK created table model dbt_demo_dwh.dim_nutrient.............. [CREATE TABLE (149.0 rows, 5.7 KB processed) in 2.94s] 14:02:54 | 5 of 8 OK created table model dbt_demo_dwh.dim_food.................. [CREATE TABLE (7.8k rows, 1.1 MB processed) in 3.38s] 14:02:56 | 7 of 8 OK created table model dbt_demo_dwh.fact_nutrition............ [CREATE TABLE (644.1k rows, 27.9 MB processed) in 5.24s] 14:02:56 | 8 of 8 START table model dbt_demo_dwh.fact_sugar_foods_top100........ [RUN] 14:03:00 | 8 of 8 OK created table model dbt_demo_dwh.fact_sugar_foods_top100... [CREATE TABLE (108.0 rows, 17.9 MB processed) in 3.73s] 14:03:00 | 14:03:00 | Finished running 4 view models, 4 table models in 11.74s. Completed successfully Done. PASS=8 WARN=0 ERROR=0 SKIP=0 TOTAL=8
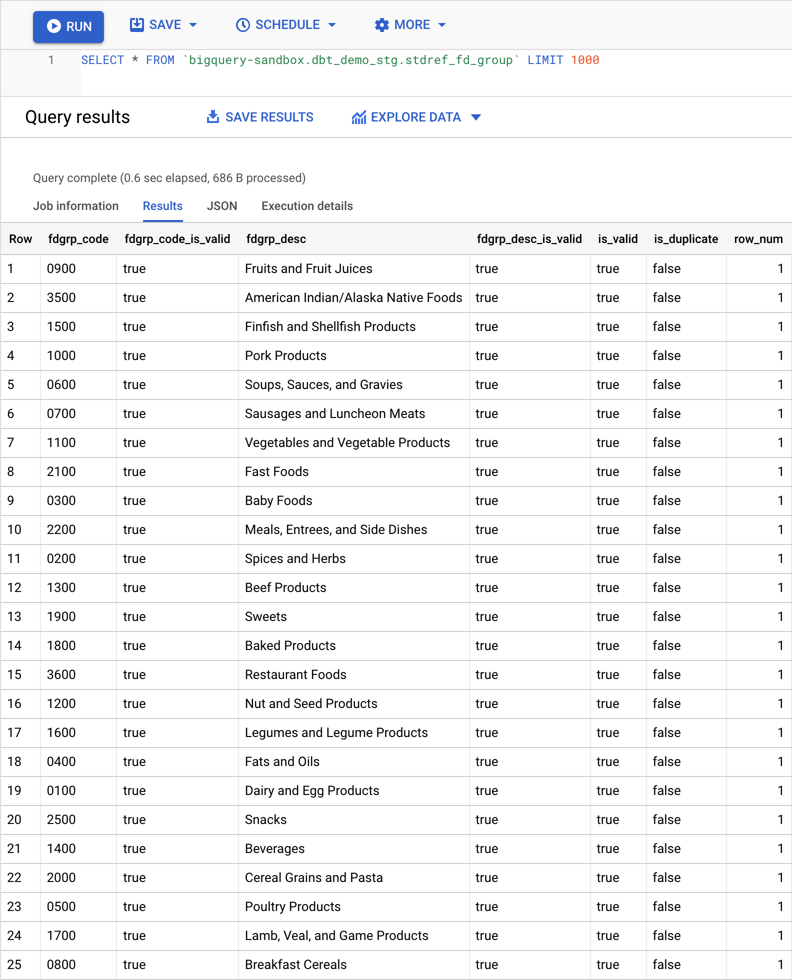
The code below shows the SQL that was generated for the stg_usda__stdref_fd_group model. It is materialized as the stdref_fd_group view in the dbt_demo_stg dataset:
with source as (
with __std_staging_query as (
select
safe_cast(left(fdgrp_cd, 4) as string) as fdgrp_code,
(safe_cast(left(fdgrp_cd, 4) as string) is not null or fdgrp_cd is null) as fdgrp_code_is_valid,
safe_cast(left(fdgrp_desc, 60) as string) as fdgrp_desc,
(safe_cast(left(fdgrp_desc, 60) as string) is not null or fdgrp_desc is null) as fdgrp_desc_is_valid,
(safe_cast(left(fdgrp_cd, 4) as string) is not null or fdgrp_cd is null) and (safe_cast(left(fdgrp_desc, 60) as string) is not null or fdgrp_desc is null) as is_valid
from `bigquery-sandbox`.`dbt_demo_ldg`.`stdref_fd_group`
)
select
*,
count(*) over(partition by fdgrp_code) > 1 as is_duplicate,
row_number() over(partition by fdgrp_code order by fdgrp_code asc) as row_num
from __std_staging_query
)
select
*
from source
Finally, below you can see the result of querying the view. As can be seen, now columns are typed, values are cleaned and extra columns were added to help identify duplicate/invalid values.
Conclusion
dbt and BigQuery can be used to standardize common data preparation steps. Adding more data sources becomes easier and faster since less code needs to be written. In effect, the dbt macros take care of generating the code based on the medata configured.
In a future post, I will show you how dbt can be integrated into a workflow. For instance, a workflow can run a series of validations after running staging models. If no data quality issues are found, then proceed to running the data warehouse models.
Thanks for reading! Please leave a comment if you have any thoughts or questions.

