Near Real-Time Data Processing for BigQuery: Part One
This post describes (near) real-time data processing for BigQuery with unique and other check constraints, and unit testing. This is part one of two, and describes the real-time ingestion of the data. Part two will describe how to implement ASSERTS on the data and unit testing inside of BigQuery.
Since December, https://blog.pythian.com/near-real-time-data-processing-for-bigquery-part-2Google has introduced new tools which allow for serverless ingestion of files and processing of data in BigQuery. These tools are:
- GCP Workflows
- Dataform
GCP Workflows
GCP Workflows were developed by Google and are fully integrated into the GCP console. They are meant to be used to orchestrate and automate Google Cloud and HTTP-based API services with serverless workflows. This means when you’re working with something which is mostly API calls to other services, Workflows is your tool of choice.
Workflows are declarative YAML. So you simply define the process you want to happen, and the workflow will take care of all the underlying effort to implement it.
Dataform
Dataform is an SaaS company that Google purchased and currently all development must still happen on their website. It’s used to develop SQL Pipelines to transform data within BigQuery without writing code.
With Dataform, you define the SQL statements you want to run. After that, they handle creating tables, views, ordering and error handling.
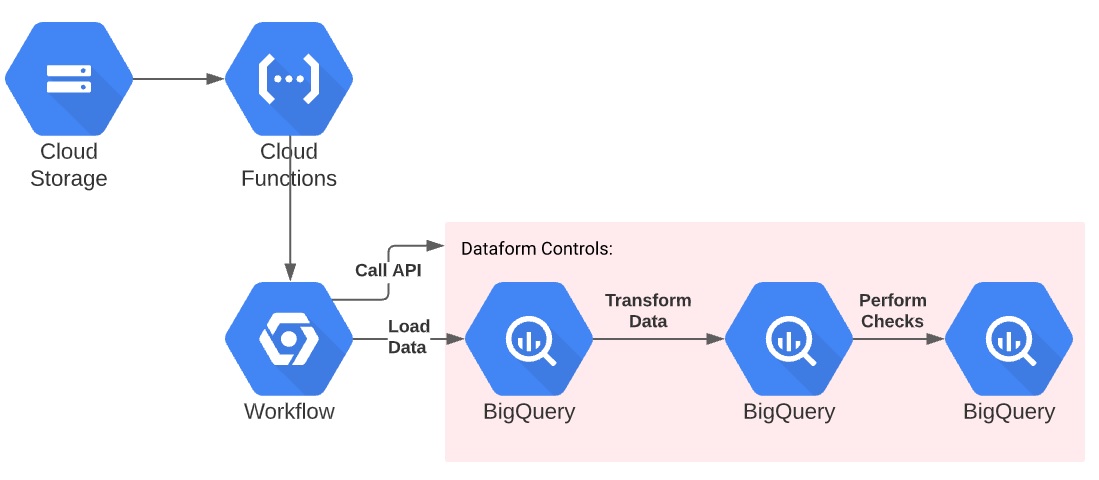
Real-time processing of flat files into BigQuery
We make use of these two tools along with existing GCP infrastructure to develop a pipeline which will immediately ingest a file into BigQuery and do all the translations needed for reporting. In addition, the pipeline will validate the data’s uniqueness and formatting. Finally, I will show how to perform unit testing of the data.
File Ingestion
The file is ingested using a GCS bucket trigger which calls the workflow. There is currently no way to trigger a workflow directly from a GCS bucket trigger.
To give proper credit, I used examples provided by Christian Kravanja and Mehdi BHA. Combining their code has produced a horrible Frankenstein which is nearly unstoppable.
This workflow:
- Accepts three parameters (bucket, file and table name).
- Starts a BigQuery load job for the file.
- Waits for the file to finish using an exponential backoff.
- Updates the file metadata to “loaded” and the load job_id.
- Returns the number of rows in the file, loaded into BigQuery, and discarded.
- And of course, logs various messages throughout.
Create GCS bucket
First things first, let’s create the infrastructure. You’ll need a GCS bucket.
Create ingestion service account
Next, we need to create a service account to run the workflow. It should have the following permissions:
- BigQuery Data Owner
- BigQuery Job User
- Logs Writer
- Storage Admin
- Workflows Invoker
Create workflow
Now, create the workflow that will start the actual load of the BigQuery job.
main:
params: [args]
#Parameters:
#
# bucket: GCS Bucket Name (no gs://)
# file: File Name. Can have wildcard
# datasetName: Dataset Name
# tableName: Table Name
#
# {"bucket":"bucket-name","file":"filename.csv","datasetName":"BQDatasetName","tableName":"BQTableName"}
# {"bucket":"staging-ingest","file":"categories.csv","datasetName":"dataform","tableName":"categories"}
steps:
# Only ten variables per assign block
- environment_vars:
# Built-in environment variables.
assign:
# Mostly not used in this code, but here to show the list of what exists
- project_id: ${sys.get_env("GOOGLE_CLOUD_PROJECT_ID")}
- project_num: ${sys.get_env("GOOGLE_CLOUD_PROJECT_NUMBER")}
- workflow_location: ${sys.get_env("GOOGLE_CLOUD_LOCATION")}
- workflow_id: ${sys.get_env("GOOGLE_CLOUD_WORKFLOW_ID")}
- workflow_revision_id: ${sys.get_env("GOOGLE_CLOUD_WORKFLOW_REVISION_ID")}
- global_vars:
# Global variables
assign:
- job_id:
- bigquery_vars:
# BigQuery job configuration
assign:
- request_body:
configuration:
load: {
destinationTable: {
datasetId: "${args.datasetName}",
projectId: "${project_id}",
tableId: "${args.tableName}"
},
sourceUris: [
"${ \"gs://\" + args.bucket + \"/\" + args.file}"
],
sourceFormat: "CSV",
autodetect: "true",
nullMarker: "NA",
createDisposition: "CREATE_IF_NEEDED",
writeDisposition: "WRITE_APPEND",
fieldDelimiter: ","
}
- log_config_state:
call: sys.log
args:
text: ${"BigQuery job configuration " + json.encode_to_string(request_body)}
severity: INFO
- load_bigquery_job:
call: http.post
args:
url: ${"https://bigquery.googleapis.com/bigquery/v2/projects/" + project_id + "/jobs"}
body:
${request_body}
headers:
Content-Type: "application/json"
auth:
type: OAuth2
result: job_response
- set_job_id:
assign:
- job_id: ${job_response.body.jobReference.jobId}
- monitor_bq_job:
try:
steps:
- get_bq_job_status:
call: http.request
args:
url: ${"https://bigquery.googleapis.com/bigquery/v2/projects/" + project_id + "/jobs/" + job_id + "?location=" + workflow_location}
method: GET
auth:
type: OAuth2
result: bq_job_status
- induce_backoff_retry_if_state_not_done:
switch:
- condition: ${bq_job_status.body.status.state != "DONE"}
raise: ${bq_job_status.body.status.state} # a workaround to pass job_state value rather that a real error
retry:
predicate: ${job_state_predicate}
max_retries: 10
backoff:
initial_delay: 1
max_delay: 60
multiplier: 2
- tag_source_object:
call: http.put
args:
url: "${\"https://storage.googleapis.com/storage/v1/b/\" + args.bucket + \"/o/\" + args.file }"
body:
metadata:
"status": "loaded"
"loadJobId": ${job_id}
headers:
Content-Type: "application/json"
auth:
type: OAuth2
- get_load_results:
steps:
- get_final_job_status:
call: http.request
args:
url: ${"https://bigquery.googleapis.com/bigquery/v2/projects/" + project_id + "/jobs/" + job_id + "?location=" + workflow_location}
method: GET
auth:
type: OAuth2
result: load_response
- log_final_job_state:
call: sys.log
args:
text: ${"BigQuery job final status " + json.encode_to_string(load_response)}
severity: INFO
- raise_error_on_failure:
switch:
- condition: ${("errorResult" in load_response.body.status)}
raise: ${load_response.body.status.errors}
- return_result:
return: >
${"Files processed: " + load_response.body.statistics.load.inputFiles +
". Rows inserted: " + load_response.body.statistics.load.outputRows +
". Bad records: " + load_response.body.statistics.load.badRecords
}
job_state_predicate:
params: [job_state]
steps:
- condition_to_retry:
switch:
- condition: ${job_state != "DONE"}
return: True # do retry
- otherwise:
return: False # stop retrying
Create cloud function
A workflow can not be currently triggered from a GCS bucket, so we need to create a cloud function to call the workflow.
So why not use the cloud function only? Well, there are a few reasons:
- Cloud Functions can only run for nine minutes, and when ingesting GB or TB of data, it’s possible this will timeout.
- Cloud Functions are pure Python, and writing even a simple API call can be complex.
- Cloud Functions are billed by duration (vs. by steps for Workflow). A long running, synchronous API call can be very expensive.
- And lastly, this wouldn’t be much of a blog post if we did that.
So, use this code to create the cloud function.
Create a requirements.txt with these values:
# Function dependencies, for example: # package>=version google-auth requests
Create a main.py file with this code:
import os.path
import json
import urllib.request
import google.auth
from google.auth.transport.requests import AuthorizedSession
def getProjectID():
url = "http://metadata.google.internal/computeMetadata/v1/project/project-id"
req = urllib.request.Request(url)
req.add_header("Metadata-Flavor", "Google")
return urllib.request.urlopen(req).read().decode()
def onNewFile(event, context):
project_id = getProjectID()
region_id = os.environ.get('WORKFLOW_REGION_ID')
tableName = event['name'].split('.')[0]
print('Event ID: {}'.format(context.event_id))
print('Event type: {}'.format(context.event_type))
print('Bucket: {}'.format(event['bucket']))
print('File: {}'.format(event['name']))
print('Dataset: {}'.format(os.environ.get('DATASET_NAME')))
print('Table: {}'.format(tableName))
scoped_credentials, project = google.auth.default(
scopes=['https://www.googleapis.com/auth/cloud-platform'])
authed_session = AuthorizedSession(scoped_credentials)
URL = 'https://workflowexecutions.googleapis.com/v1/projects/{}/locations/{}/workflows/bigquery-fileload/executions'.format(project_id, region_id)
params_dict = { \
'bucket': '{}'.format(event['bucket']),
'file': '{}'.format(event['name']), \
'datasetName': '{}'.format(os.environ.get('DATASET_NAME')), \
'tableName': '{}'.format(tableName) }
PARAMS = { 'argument' : json.dumps(params_dict) }
response = authed_session.post(url=URL, json=PARAMS)
print(response)
And finally, deploy the cloud function using this script:
gcloud functions deploy loadfiletobigquery \ --region [GCP-REGION] \ --entry-point onNewFile \ --runtime python38 \ --set-env-vars WORKFLOW_REGION_ID=[WORKFLOW_REGION_ID],DATASET_NAME=[BQ_DATASET_NAME] \ --trigger-resource [GCS-BUCKET-NAME] \ --trigger-event google.storage.object.finalize \ --service-account [Ingestion SERVICE-ACCOUNT-NAME]
End of part one
Now we have an ingestion process setup that will take any flat file and load it into BigQuery. Just place the file on the GCS bucket, wait a few seconds, and you’ll see it automagically appear.